IBM to Study Human Microbiome’s Role In Autoimmune Disorders.
The microbiome is emerging as a new player in human health. IBM just announced plans to study the human microbiome and its role in autoimmune diseases. [This article first appeared on the website LongevityFacts.com. Author: Brady Hartman. ]
IBM just announced plans to study the human microbiome and its role in autoimmune diseases.
Despite a major research initiative by the U.S. Government, we still don’t fully understand how the bacteria inside us affects our health. IBM plans to find out more by analyzing millions of microbial genes, starting with those belonging to gut bacteria. The hope is that this could shed light on conditions such as Crohn’s disease, Hashimoto’s, type 1 diabetes, and ulcerative colitis.
To increase resources on the project, IBM is crowd-sourcing extra computing power through its World Community Grid, asking anyone with a computer to help.
Scientists from several US universities will oversee the analysis, with the goal of finding new ways to prevent or treat autoimmune diseases. For medical expertise on the study, IBM has partnered up with the University of California San Diego, the Broad Institute of Harvard and MIT, the Simons Foundation’s Flatiron Institute, and Massachusetts General Hospital. IBM aims to map three million bacterial genes found in the human microbiome.
IBM isn’t the only tech firm setting its sights on the microbiome. In fact, it seems to be the latest vogue for tech companies. In April, Google’s life sciences arm Verily launched a project aiming to collect microbiome and genetic data from 10,000 people in the US. Their goal is to better predict the onset of conditions like cancer and heart disease. Tech billionaire Naveen Jain began his own exploration of the nascent field with the creation of startup firm Viome in 2016. Google is investing heavily in the life sciences. Four years ago, the tech giant launched Calico Labs, a $1.5 Billion dollar startup with the sole mission of curing the diseases of aging. According to the Wall Street Journal, in the past couple of years, several startups have been gobbling up venture capital to study the subject.
IBM will leverage the groundwork laid by the Human Microbiome Project. The NIH’s 5-year, $115 million dollar project has already mapped the human microbiome using genome sequencing techniques. Human Microbiome Project researchers have made IBM’s job easier by creating a reference database of normal microbial variation in humans.
The Microbiome’s Role In Human Health
The microbes that colonize the surface and insides of our bodies are essential for life. We are dependent on these bacteria to produce certain vitamins and regulate our immune system. The tiny critters in our microbiome help digest our food and keep us healthy by protecting us against disease-causing bacteria.
It’s a pretty large ecosystem. Microbes in a human adult are estimated to outnumber human cells by a ratio of ten to one. Even though microbes are only one-tenth to one-hundredth the size of a human cell, the microbiome as a whole may account for up to 2 kilos (five pounds) of adult body weight.
The U.S. Government and the Microbiome
The microbiome is so important that the feds spent millions of dollars on a 5-year study called the Human Microbiome Project. This unparalleled study has provided us more insight into the human microbiome in the last several years than in the prior 50 years combined.
The Human Microbiome Project
The Human Microbiome Project is an initiative of the National Institutes of Health (NIH). Launched in 2008, the revolutionary project mapped out the human microbiome in all its complexity. The goal of the project’s sponsors is to identify and characterize flora and fauna in the human microbiome, including those in both healthy and diseased humans. The project was best characterized as a feasibility study and had a total budget of $115 million. Before the project, the microbiome was poorly understood. While the NIH has sponsored similar projects investigating the microbiome, the ultimate goal of the project was to test how changes in the human microbiome are associated with human health or disease.
Mapping the Human Microbiome
Members of the Human Microbiome Project team obtained samples from 242 healthy volunteers, collecting more than 5,000 samples from over 15 bodily sites such as the nose, mouth, skin, lower intestines, and vagina. The researchers analyzed the human and microbial DNA with DNA sequencing machines. Team members extracted microbial genomic data by identifying the bacterial specific ribosomal RNA, called “16S rRNA”. The research team calculated that more than 10,000 microbial species occupy the human microbiome and also identified 81 to 99% of the genera.
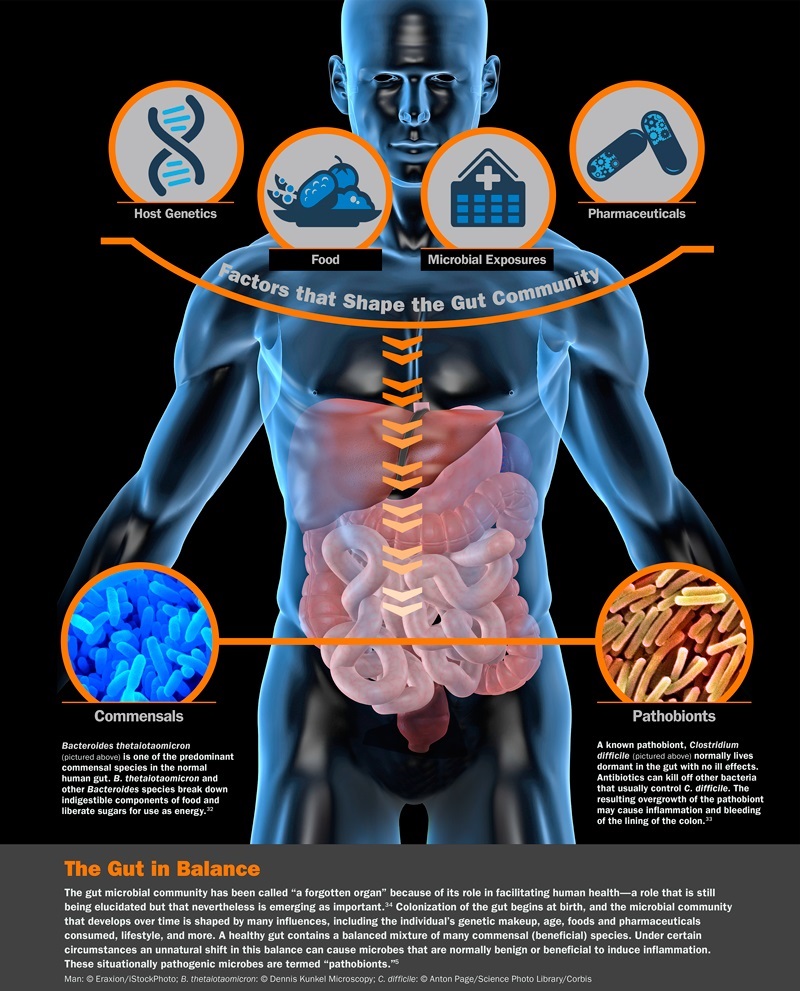
Key Microbiome Project Components
Key components of the project included culture-independent methods of microbial community characterization, such as metagenomics and whole genome sequencing. The metagenomics aspect provided a broad genetic perspective on a single microbial community, while the entire genome sequencing provided a deep genetic perspective on particular aspects of a given microbial community, such as an individual bacterial species. The latter served as reference genomic sequences for comparison purposes during subsequent metagenomic analysis. To map out the microbiome, the researchers planned about 3000 genomic sequences of individual bacterial isolates. The project studied the microbial ecosystem of five bodily sites, including the gut, mouth, skin, vagina, lung and nasal passages. Using samples from human subjects, the project team also performed deep sequencing of bacterial 16S rRNA sequences amplified by polymerase chain reaction.
Human Microbiome Project Achievements
Team members didn’t just produce time-lapse moving pictures of the human microbiome; they also published over 190 peer-reviewed papers. Team members primarily identified factors distinguishing the microbiota of healthy and diseased gut. They also developed new predictive methods for identifying active transcription factor binding sites. Team members identified a hitherto unrecognized dominant role of Verrucomicrobia in soil bacterial communities. Using bioinformatic evidence, the microbiome researchers identified a widely distributed, ribosomally produced electron carrier precursor. The members of the research team identified unique adaptations adopted by segmented filamentous bacteria in their role as gut commensals. These bacterial residents of the microbiome are medically important because these tiny critters stimulate T helper 17 cells, thought to play a key role in autoimmune disease. The team members identified factors determining the virulence potential of strains of the bacteria Gardnerella vaginalis that causes vaginosis. The microbiome researchers identified a link between oral microbiota and atherosclerosis. And finally, team members demonstrated that pathogenic species of Neisseria – a bacteria involved in meningitis, septicemia, and sexually transmitted diseases – exchanges virulence factors with commensal species.
NIH Director Francis Collins announced a significant milestone in 2012. By mapping the human microbiome using genome sequencing techniques, project researchers had created a reference database of normal microbial variation in humans.
Clinical Uses of the Human Microbiome Project
Among the first clinical applications utilizing data from the Human Microbiome Project, researchers found a shift to less species diversity in the vaginal microbiome of pregnant women before birth. As well, the research team found a high viral DNA load in the nasal microbiome of children with unexplained fevers. Other studies using data from the project have gained a better understanding of the role of the microbiome in various childhood disorders as well as diseases of the skin, reproductive organs, and digestive tract.
Summary Of Microbiome Research
In addition to constructing the first human microbiome reference database, members of the research team also discovered several surprises, which include:
- The massive ecosystem contributes more genetic code responsible for human survival than our genes. Team members estimated that bacterial protein-coding genes in the human microbiome are 360 times more abundant than human genes.
- Microbial metabolic activities in the healthy microbiome – such as the digestion of fats – are not always provided by the same bacterial species.
- The researchers found out that the microbiome changes over time and it’s affected by medications you take and the diseases that you have. For example, people with type 2 diabetes have a different microbiome than those without the condition.
- Even a healthy microbiome changes when you have an infection, such as a cold or fever. However, the microbiome eventually returns to a state of equilibrium.
- Most notably, researchers found that the diversity of bacteria in our microbial ecosystem benefits human health. A healthy microbiome is a healthy body.
Bottom Line
The human microbiome is becoming an emerging area of anti-aging research. However, scientists still have much to learn about our microbial ecosystem.
Related Articles
- Some people are altering their microbiome to eradicate a bad bug and getting fecal transplants by swallowing pills made out of medical poop.
- Researchers recently extended the lifespan of middle-aged animals by nearly 50% with a fecal transplant, showing that the poop of younger fish rejuvenated the older ones by upgrading their microbiome.
- Excellent introductory video to the role of the microbiome in human health – How Bacteria Rule Over Your Body – the Microbiome.
- Scientists have linked changes in the microbiome to chronic low-grade inflammation, known as inflammaging.
- Researchers show that ‘ridiculously healthy’ elderly have the same gut bacteria composition as healthy 30 year-olds in microbiome study of centenarians.
Help Us Carry the Message
Please share this post and help us spread the word. All it takes is one simple click on any social media link on this page.
Disclaimer
Diagnosis, Advice, and Treatment: This article is intended for informational and educational purposes only and is not a substitute for professional medical advice. The information provided in this report should not be used during any medical emergency or for the diagnosis or treatment of any medical condition. Consult a licensed physician for the diagnosis and treatment of any and all medical conditions. Call 911, or the equivalent emergency hotline number, for all medical emergencies. As well, consult a licensed physician before changing your diet, supplement or exercise programs.
Photos, External Links & Endorsements: This article is not intended to endorse companies, organizations or products. Links to external websites, depiction/mention of company names or brands, are intended only for illustration and do not constitute endorsements.
Very good post. Thank you
Just one suggestion… the pictures are off kilter on my tablet
Nice post about the tech firms
Well written, thanks